Akhil KumarI am a Chemical Engineering undergraduate from Indian Institute of Technology (IIT) Delhi. I am an Associate Researcher in Tsankov Lab at the Icahn School of Medicine at Mount Sinai, where I leverage single-cell genomics approaches to advance precision medicine for colorectal cancer. Before this, I was a Research Fellow in Perumal Lab at the School of Biological Sciences of Indian Institute of Technology (IIT) Delhi, where I used evolutionary bioinformatics approaches to better understand host adaptation dynamics of SARS-CoV-2 and Hepatitis C virus. Email / GitHub / Google Scholar / LinkedIn / CV |

|
Published ResearchI'm interested in addressing biomedical problems, particularly in cancer, through functional genomics and systems biology, with an emphasis on computational method development. Selected works are highlighted. |
|
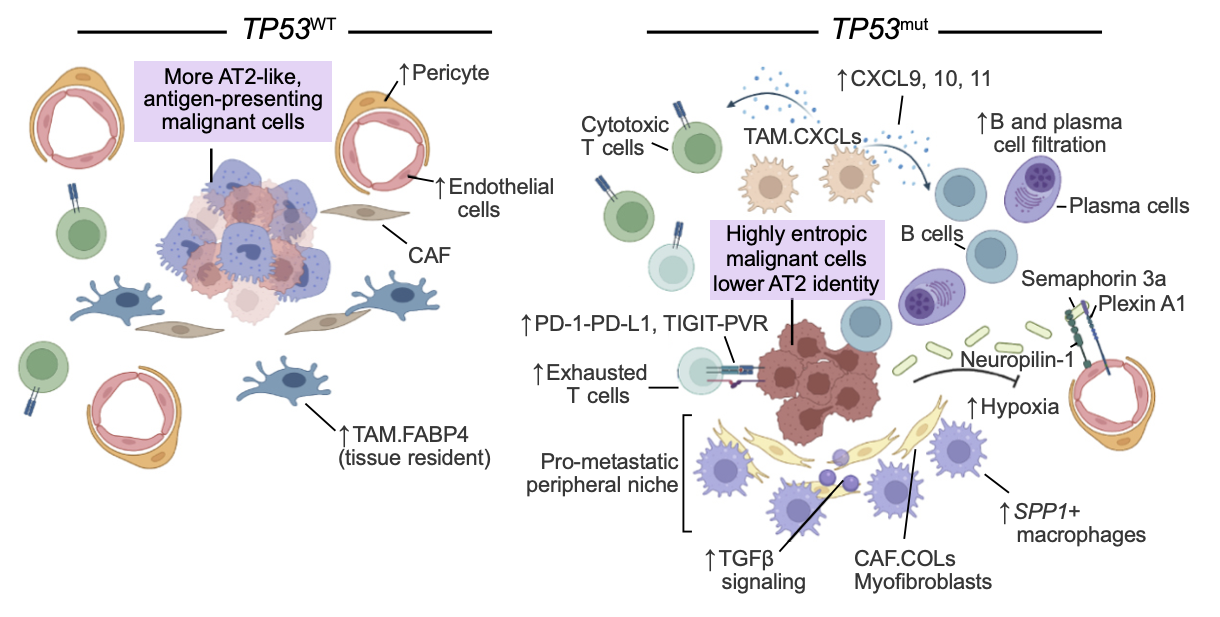
A cellular and spatial atlas of TP53-associated tissue remodeling in lung adenocarcinoma.William Zhao*, Thinh T Nguyen*, Atharva Bhagwat*, Akhil Kumar*, Bruno Giotti, ..., Aaron N Hata , Aviv Regev , Bruce E Johnson , Alexander M Tsankov Nature Cancer, 2025 paper / code / We constructed a multiomic cellular and spatial atlas of 23 treatment-naive human lung tumors to define how TP53 mutations affect the Lung Adenocarcinoma tumor microenvironment. My work involved validating trends observed in single-cell data across two independent bulk cohorts. |
|
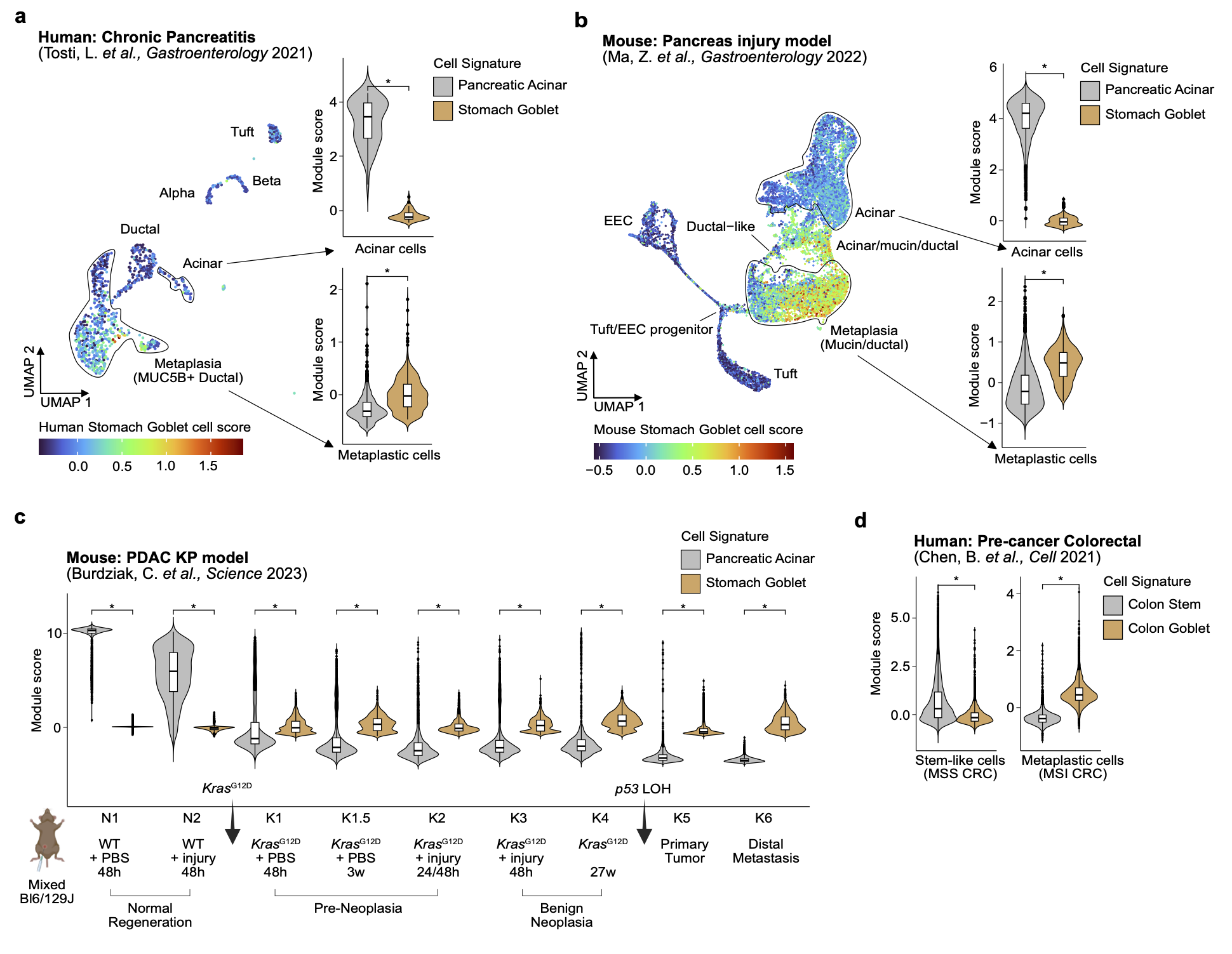
Cancer Cell of Origin.Mohamad D Bairakdar*, Wooseung Lee*, Bruno Giotti*, Akhil Kumar*, Paula Stancl*, Elvin Wagenblast, Dolores Hambardzumyan, Paz Polak, Rosa Karlic, Alexander M Tsankov Nature communications, 2025 paper / code / We combined 3,669 whole genome sequencing patient samples, 559 single-cell chromatin accessibility cellular profiles, and machine learning (ML) to predict the COO of 37 cancer subtypes with high robustness and accuracy, confirming both the known anatomical and cellular origins of numerous cancers, often at cell subset resolution. My work validated the hypothesis, generated using our ML prediction model, of an intermediate metaplastic state during tumorigenesis for multiple gastrointestinal cancers, which have important implications for cancer prevention, early detection, and treatment stratification. |
|
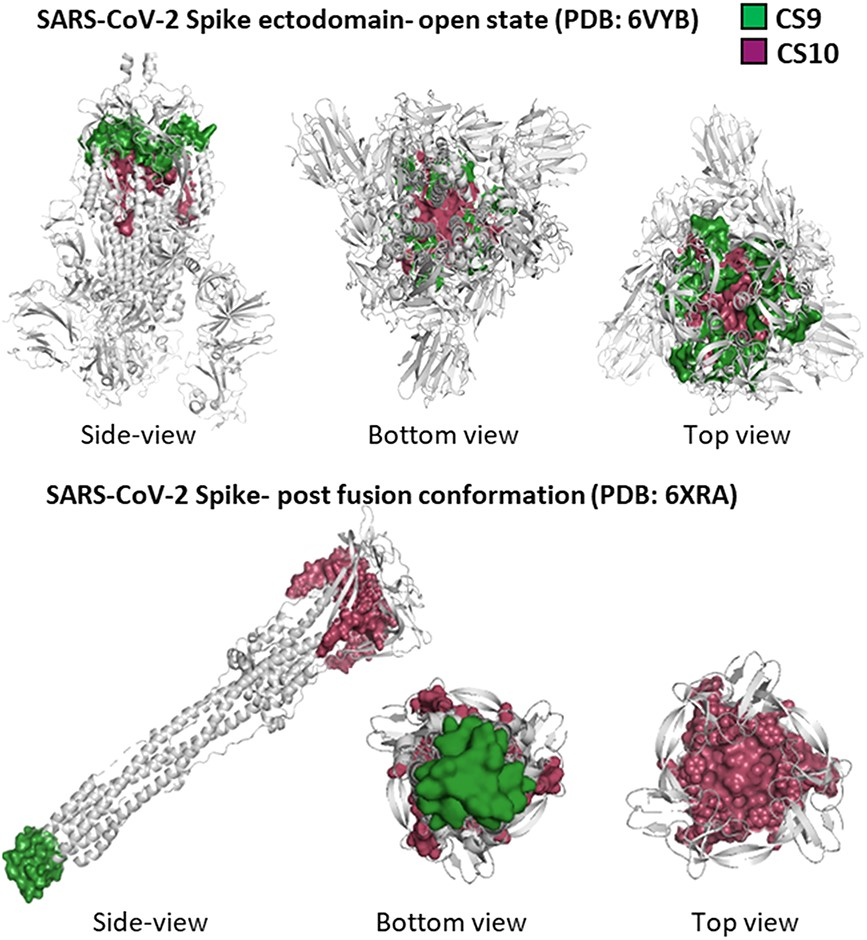
Improvement of diagnostic assays against emerging variants of SARS-CoV-2.Akhil Kumar, Rishika Kaushal, Himanshi Sharma, Khushboo Sharma, Manoj B Menon, Vivekanandan Perumal Briefings in Functional Genomics, 2023 paper / code / We identified 11 conserved stretches in over 6.3 million SARS-CoV-2 genomes including all the major variants of concerns. Each conserved stretch is ≥100 nucleotides in length with ≥99.9% conservation at each nucleotide position. Interestingly, six of the eight conserved stretches in ORF1ab overlapped significantly with well-folded experimentally verified RNA secondary structures. Our findings highlight the role of structural constraints at both RNA and protein levels that contribute to the sequence conservation of specific genomic regions in SARS-CoV-2. |
|
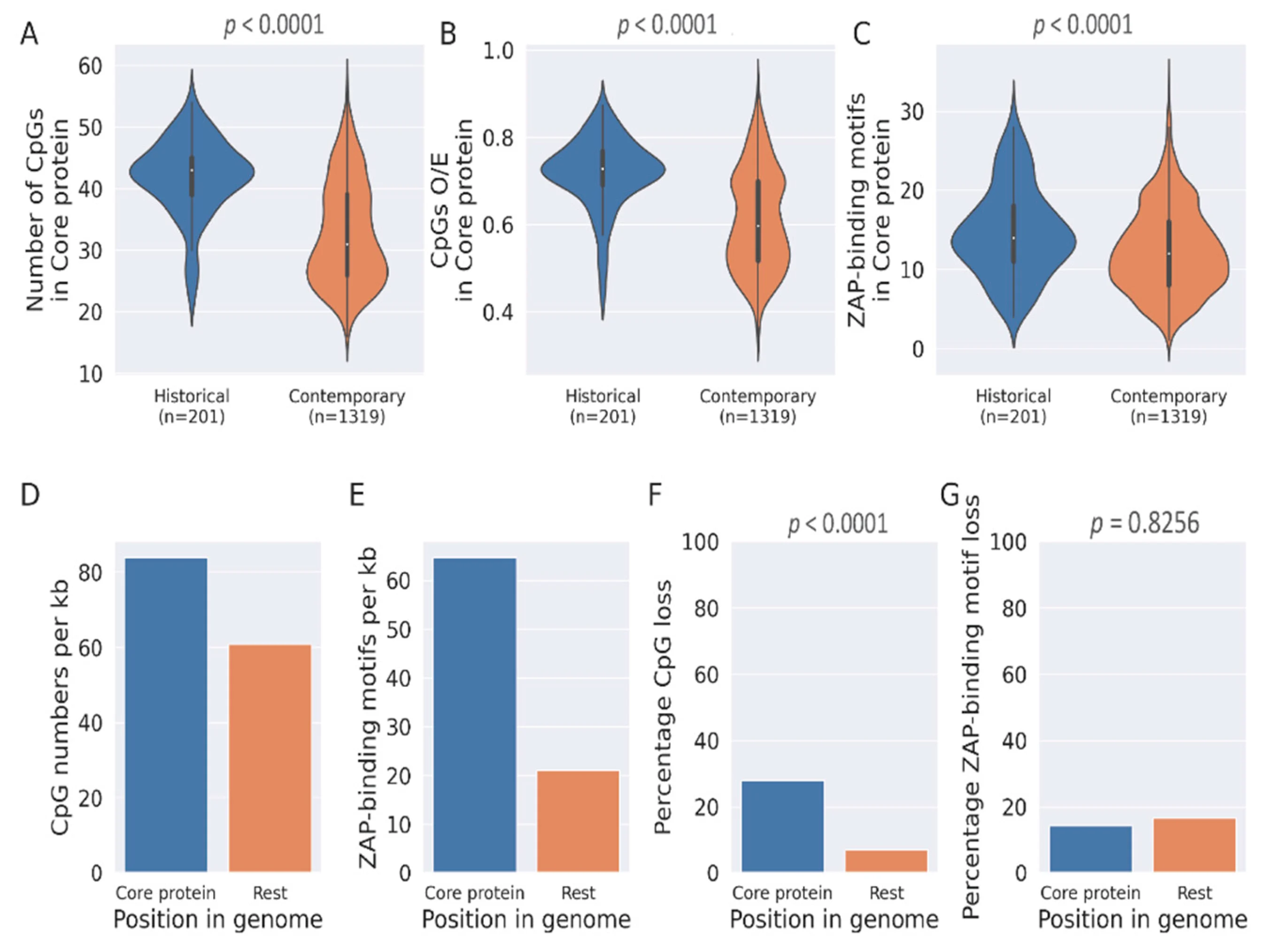
Host-adaptation dynamics in Hepatitis C virus.Sanket Mukherjee*, Akhil Kumar*, Jasmine Samal, Ekta Gupta, Vivekanandan Perumal, Manoj B Menon Pathogens, 2022 paper / code / This work highlights CpG depletion in HCV genomes during their evolution in humans and the role of ZAP-mediated selection in HCV evolution. |
|
Host-adaptation dynamics in SARS-CoV-2.Akhil Kumar*, Nishank Goyal*, Nandhini Saranathan, Sonam Dhamija, Saurabh Saraswat, Manoj B Menon, Vivekanandan Perumal Molecular Biology and Evolution, 2022 paper / code / This work highlights how temporal variations in selection pressures during virus adaption may impact the rate and the extent of CpG depletion in virus genomes. |
Work Under ReviewRecent findings currently submitted for peer review. |
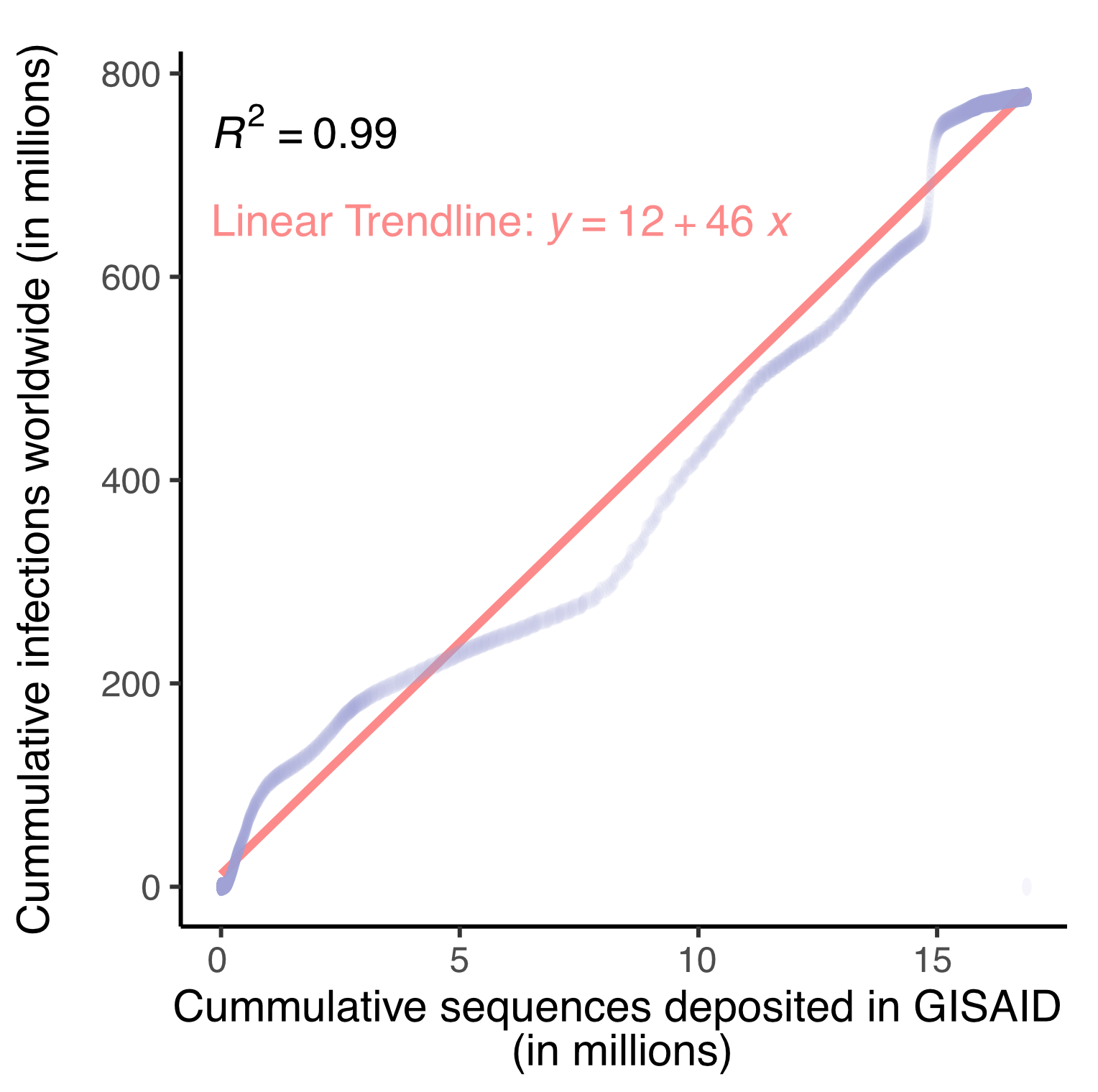
|
Early mutation blitzkrieg in SARS-CoV-2.Akhil Kumar*, Rishika Kaushal*, Manoj B Menon, Vivekanandan Perumal Under-review, 2025 code / Analysis of 16.7 million SARS-CoV-2 genomes to understand mutation emergence patterns during viral evolution. |
Additional ResearchA collection of prior projects. |
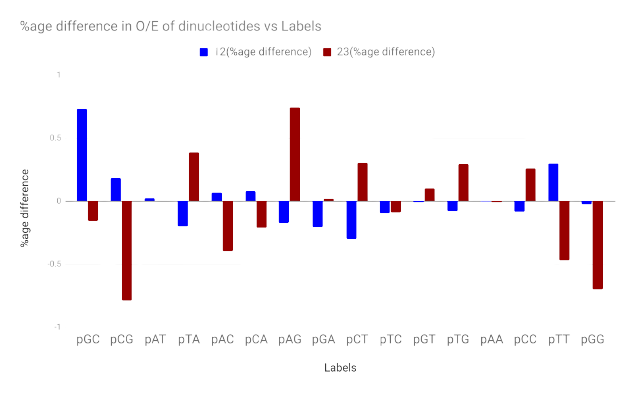
|
Temporal evolution of mono- and dinucleotides in human mitochondrial DNAPerumal Lab, Indian Institute of Technology Delhi Independent study 2019-11-28 slides / Inspected temporal evolution of mono- and dinucleotides in human mtDNA by analysing sequences dated back to 50k BC. Developed boxplots for samples from earliest-, middle- and latest-period; conducted statistical tests to compare their medians. Our findings suggested that lighter strand is becoming heavier, and a slight bias toward GC-rich sequences in ancient reads. |
|
In-silico design of small ligand molecules to inhibit early-stage insulin aggregation nucleationMultiscale Modeling Group, Indian Institute of Technology Delhi Coursework 2019-11-24 slides / Assisted with the in-silico design of small ligand molecules to inhibit early-stage insulin aggregation nucleation. Identified EGCG and polyoxometalates as the putative ligands through extensive literature review, programmed their structures. Performed targeted docking of EGCG into a partially folded intermediate of insulin using Gray lab’s ROSIE server. |
|
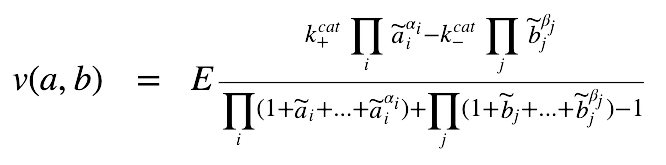
Systematic construction and validation of kinetic model from metabolic networksBiomolecular Computational Group, Indian Institute of Science Bangalore Summer Internship 2019-07-15 slides / Investigated existing kinetic modelling approaches to translate metabolic networks of interest into a dynamic model. Designed a convenience kinetics based dynamic model comprising 92 reactions, 110 species using complex pathway simulator. Distributed the reactions across 4 compartments and simulated its dynamic characterstics; examined robustness of the model. |
|
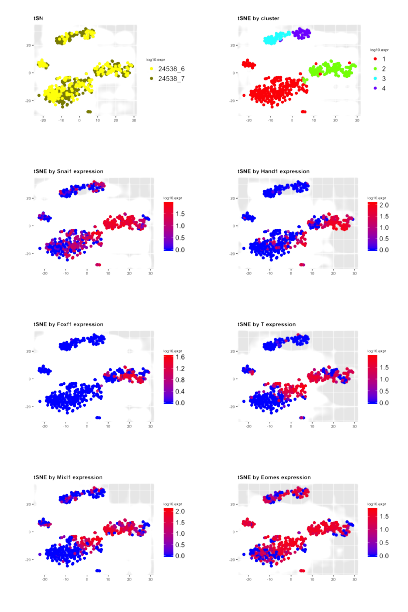
Single-cell transcriptomic data analysisSrinivas Group, University of Oxford Summer Internship 2018-07-24 code / Analysed single-cell transcriptomic data collected from early mouse embryo mutant carrying a mutation in ASPP-2 gene. Processed sequence data files, mapped sequence reads, performed quality control on the individual cells, normalized the data. Identified cell sub-types using an unbiased hierarchical clustering algorithm and expression values for biological marker genes.
|
BlogComing soon. |
|
Design and source code from Leonid Keselman's Jekyll fork of Jon Barron's website. |